Essential Gene Knowledge Graph
Citation: Systematic identification of cancer-type-specific drugs based on essential genes and validations in lung adenocarcinoma.
Lian X, Kuang X, Zhang DD, Xu Q, Ye A, Wang CY, Cui HT, Guo HX, Zhang JY, Liu Y, Hao GF, Cheng Z, Guo FB.
Brief Bioinform. 2025 May 1;26(3):bbaf266. doi: 10.1093/bib/bbaf266.
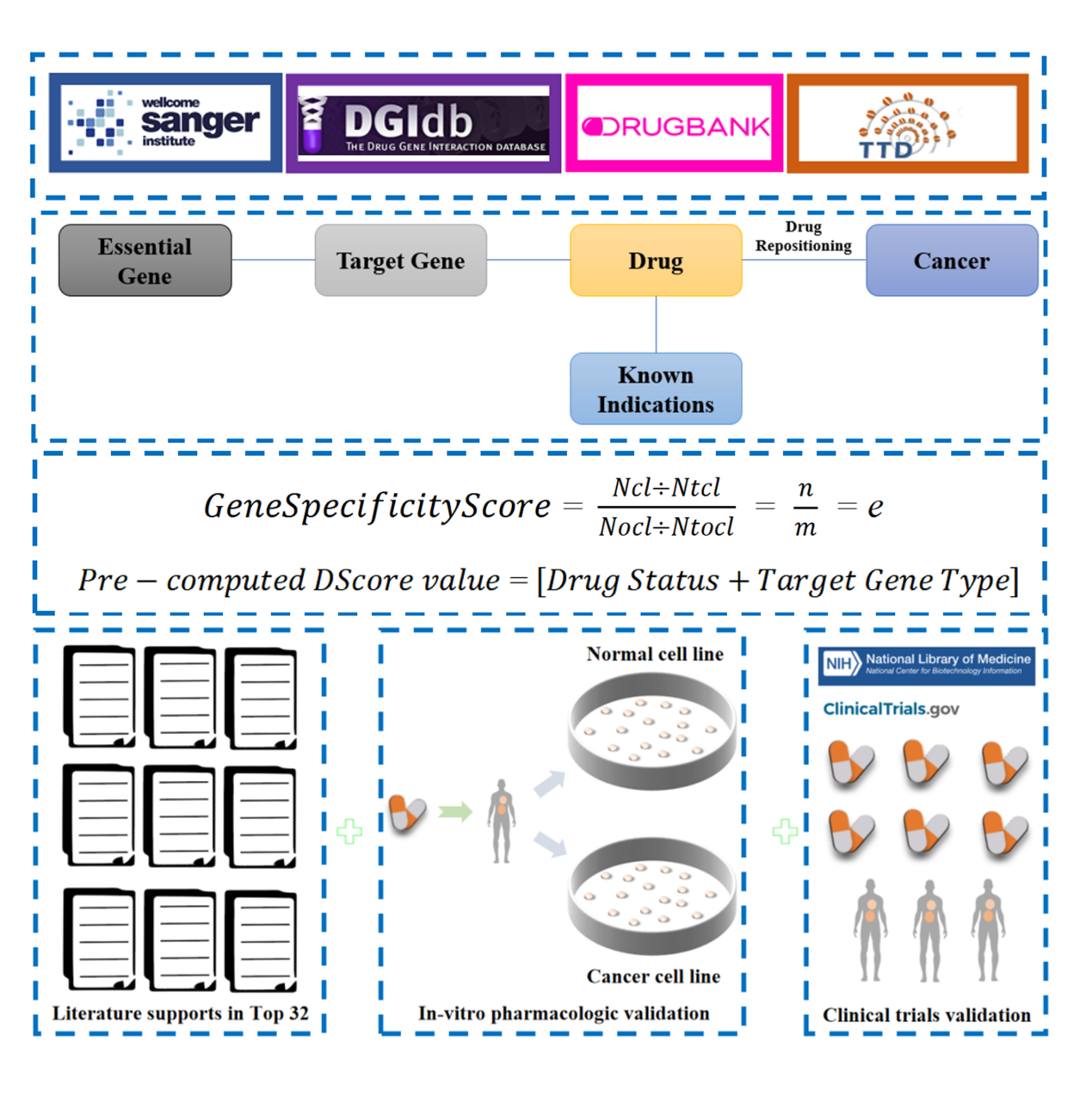
Based on specificity of gene essentiality in various cancer types, we developed a new strategy to screen targets and identify candidate drugs. Specifically, we defined GeneSpecificityScore (e score) for reflecting the specific essentiality of genes in cancer types. We then ranked the genes by GeneSpecificityScore and selected the top ones as drug targets. Secondly, combining target-drug interaction databases TTD (https://db.idrblab.net/ttd/) and DGIdb (https://www.dgidb.org/) with the research/marketing status of existing drugs/compounds, potential drugs against cancer-type-specific targets were picked out.
We verified the feasibility of our strategy through experiment. All of the seven identified drugs with no prior anti-cancer evidences were validated through inhibiting experiment in 11 cell lines of lung adenocarcinoma. Furthermore, lower inhibiting rates in 10 normal cell lines were observed for all the drugs. Colchicine was found to have the highest inhibiting effect on lung adenocarcinoma cells. We also proposed the concept of orthogonal drug targets to summate the effect of multiple drugs and verified the anti-lung cancer effects of progesterone and rosiglitazone in vitro. The results of cell level showed that using both drugs simultaneously could achieve improved chemotherapy effects while greatly reducing the damage to normal cells. Our proposed systematic strategy could be used to screen type-specific targets and find more therapies for other cancers.
The online service was established to graphicly display the relationship between cancer types, essential genes, and drugs. In addition, the results of our experiments were graphically displayed. And for external validation, the results of cellular experiments from DepMap were linked to.