Overall of the GMMAD2
GMMAD2: a comprehensive database of human gut microbial metabolite-disease associations
The natural products, metabolites, of gut microbe are the crucial effect factors on diseases. Comprehensive identification and annotation of relationship among diseases microbe metabolites can provide accurate and targeted solutions towards understanding the mechanism of complex disease and development of new markers and drugs.
To help researchers better understand the mechanisms of metabolite regulation and find potential new drugs, we built the GMMAD2 (Gut Microbial Metabolite Association with Disease, http://gepa.org.cn/GMMAD2) which aims to provide users comprehensive metabolite related information of gut microbe in human diseases.
Currently, GMMAD2 is free to access, browse, search, and download.
Help
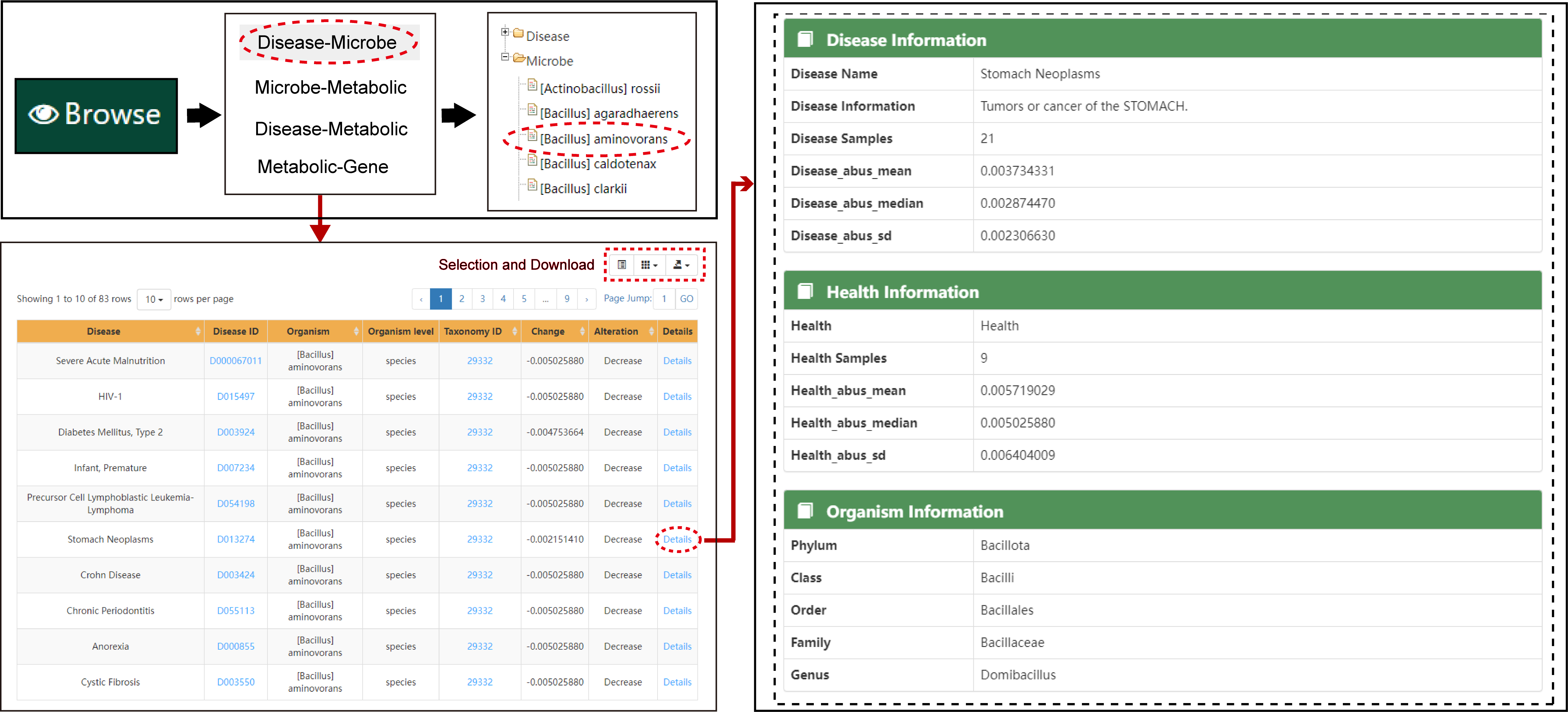
Browse Page
The browse page provides users with three options: browse by Disease-Metabolite, Disease-Microbe or Microbe-Metabolite. The disease-microbe associations were collected from GMrepo. The microbe-metabolite associations were obtained from VMH, gutMGene, Web of Microbes and Metabolomics Data Explorer. The disease-metabolite association were predicted by our method. Users can select the entries under the categories of interest to view specific information. And, the browse page allows the user to download the browsing table.
The following figure shows the flow of browsing associations.
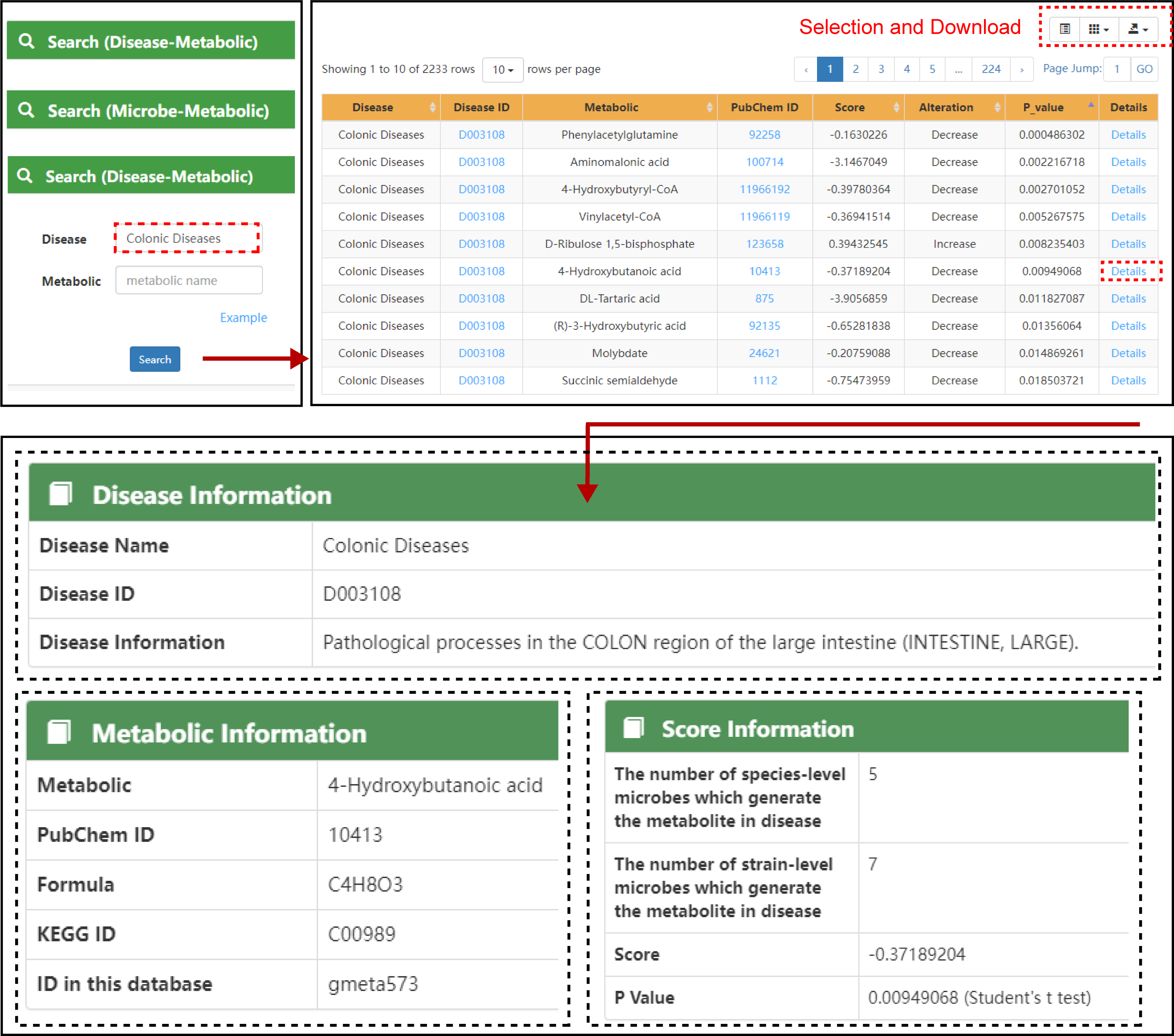
Search Page
In this interface, users can input disease name, microbe species name or metabolites name to obtain the association information. GMMAD2 also provides examples to give users a clearer understanding of how to search.
The following figure shows the flow of searching associations.
Download Page
The Download page provides file downloads of all entries in GMMAD2 which records Disease-Metabolite, Disease-Microbe or Microbe-Metabolite relationships.
Meanwhile, it also provides external links to relevant website.
Statistics Page
The statisics page provides the number of kinds of entries in GMMAD2.
Association Score & Confidence Score
We defined an association score to predict the association between the diseases and metabolites. We define the abundance of gut microbiota at the strain level in patients as D. Similarly, the abundance of gut microbiota at the strain level in healthy individuals is defined as H. These abundances are assumed to be numerically equivalent to species abundance.
The formula is shown below:
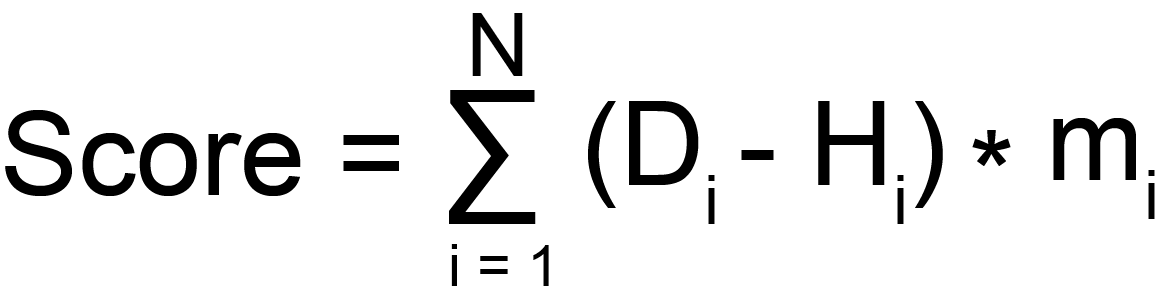
The above formula, mi represents whether the gut microbes in this disease produces this metabolite. In addition, we selected microbes with sequencing samples greater than 4 and ranked among the top 100 in disease vs health abundance changes to calculate the score.

Meanwhile, we use paired sample t-tests to calculate confidence levels between the disease and metabolic.
The formula is shown below:
